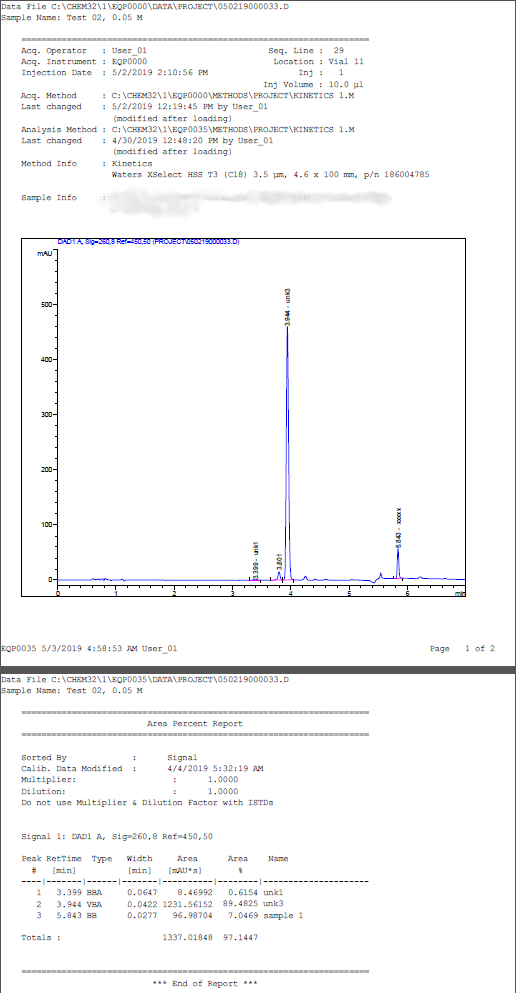
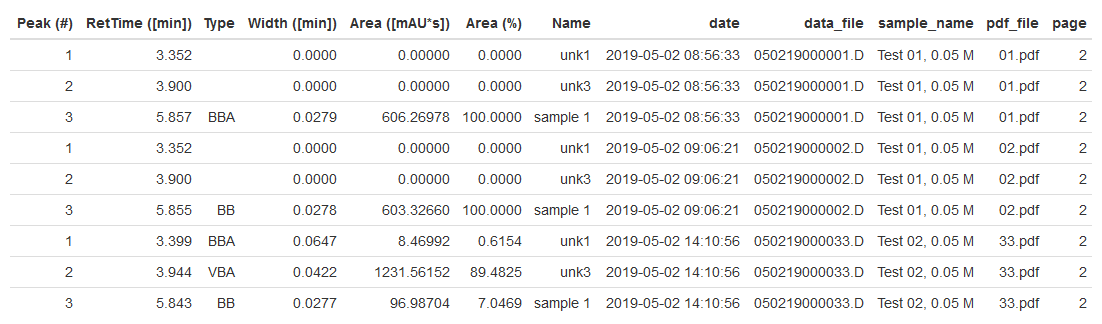
Extracting data from LCMS PDF
Here's an example of using R to extract tabular content and metadata from a series of pdf files and push the data into a single table. The concept is fairly straightforward - read in PDF and convert to text, parse each line and use regular expressions to extract pertinent information.
Three types of data are returned:
- peak data. Peak number, retention time, type, width, area and name. In fact all columns will be extracted from the table along with the column identifiers and units.
- metadata. In this case injection date, data file name and sample name are pulled from the pdfs.
- file data. The pdf file name and page containing the peak table.
The pdftools library is used to read in the PDF files.
Unwanted or Missing spaces
So why use column identifiers from the table header instead of just splitting table columns using a delimeter such as a tab? Unfortunately tabular information is lost when the pdf is imported and any tabs are converted into spaces. A space character column delimeter could be used if all columns are populated and the sample names did not contain spaces. For these tables, however, the Type column is not always populated and the Name column can contain spaces. Therefore there is no way to be sure that we are accurately identifying the end of one column and beginning of another using delimeters.
An additional issue arises because when the pdf is imported some unexpected formatting issues can arise, particularly with respect to unwanted or missing spaces. To account for this when parsing the table we can nudge forward or back to identify spaces between values. This can be seen in the alignment of line 3 of the table below (pdf vs parsed text).
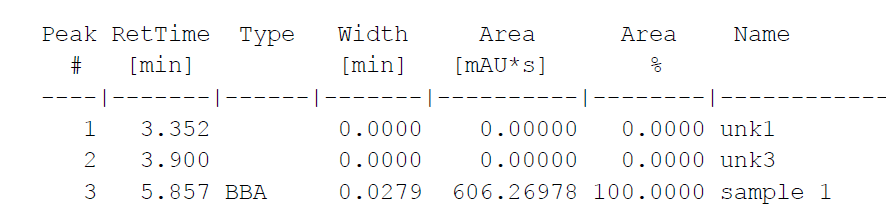
PDF Version
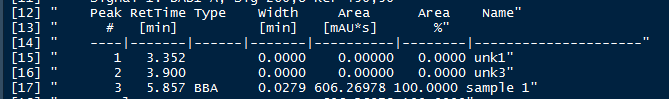
Parsed Version
Code
1## table_extract
2## extract table contents along with headers
3## this allows sample names to be retrieved too
4## uses heading underline separators to determine column widths
5
6library(pdftools)
7library(stringr)
8library(dplyr)
9
10folder <- 'test_01'
11
12## identify files
13f_names <- list.files(path = folder, pattern = '.pdf', full.names = TRUE)
14
15## loop over all files
16l.df <- lapply(f_names, function(f) {
17
18 txt <- pdf_text(f)
19
20 ## split text at carriage return
21 txt1 <- strsplit(txt, '\r\n')
22
23 ## loop over lines and retrieve table contents
24 l <- list()
25 l_all <- list()
26 read_table <- FALSE
27 for (i in 1:length(txt1)) { # loop through pages
28 for (j in 1:length(txt1[[i]])) { # loop through lines
29 line <- txt1[[i]][j]
30
31 if (str_detect(line, 'Totals :')) { # end of record
32 read_table <- FALSE
33 df.data <- as.data.frame(do.call(rbind, l_data), stringsAsFactors = FALSE)
34 df.data$date <- l[['date']]
35 df.data$data_file <- l[['data_file']]
36 df.data$sample_name <- l[['sample_name']]
37 df.data$pdf_file <- basename(f)
38 df.data$page <- i
39 l[['data']] <- df.data
40 l_all[[length(l_all) + 1]] <- l
41 l <- list() ## reset list
42 } else if (str_detect(line, 'Injection Date')) { # found inj date
43 inj_date <- as.POSIXct(str_extract(line, '\\d+/\\d+/\\d+ \\d+:\\d+:\\d+ [A|P]M'), format = '%m/%d/%Y %I:%M:%S %p')
44 l$date <- inj_date
45 l_data <- list()
46 } else if (str_detect(line, 'Peak RetTime')) { # found table
47 table_headers <- line ## save table headers for later use
48 read_table <- TRUE
49 } else if (str_detect(line, 'Data File')) {
50 l[['data_file']] <- basename(txt1[[2]][1])
51 } else if (str_detect(line, 'Sample Name:')) {
52 l[['sample_name']] <- str_replace(line, 'Sample Name: ', '')
53 }
54
55 if (read_table) {
56
57 if (str_detect(line, '\\[min\\]')) { # found second header line
58 table_headers_2 <- line ## save table headers for later use
59 }
60
61 if (!str_detect(line, '\\[min\\]') & !str_detect(line, 'Peak RetTime')) {
62 if (str_detect(line, '--|--')) { ## found separator line
63 locate_separators <- str_locate_all(line, '\\|')
64 locate_separators <- c(1, locate_separators[[1]][,1], nchar(line))
65 txt_headings <- sapply(seq_along(locate_separators[-1]), function(x) {
66 trimws(substring(table_headers, locate_separators[x], locate_separators[x+1]))
67 })
68
69 txt_headings_2 <- sapply(seq_along(locate_separators[-1]), function(x) {
70 trimws(substring(table_headers_2, locate_separators[x], locate_separators[x+1]))
71 })
72
73 txt_headings <- sapply(seq_along(txt_headings), function(x) {
74 if (nchar(txt_headings_2[x]) > 0) {
75 sprintf('%s (%s)', txt_headings[x], txt_headings_2[x])
76 } else {
77 txt_headings[x]
78 }
79 })
80
81 } else { ## found data
82
83 ## add a space to the end of the line
84 ## necessary for identifying end-of-data points
85 line <- paste0(line, ' ')
86
87 data <- sapply(seq_along(locate_separators[-1]), function(x) {
88
89 ## sometimes text pull needs to start a few characters back
90 ## (due to import conversion of tabs to spaces)
91 start_pos <- locate_separators[x]
92 end_pos <- locate_separators[x+1]
93
94 if (substring(line, start_pos, start_pos) != ' ') {
95 found_space <- FALSE
96 while (!found_space & start_pos > 2) {
97 start_pos <- start_pos - 1
98 if (substring(line, start_pos, start_pos) == ' ') found_space <- TRUE
99 }
100 }
101
102 if (substring(line, end_pos, end_pos) != ' ') {
103 found_space <- FALSE
104 while (!found_space & end_pos > 2) {
105 end_pos <- end_pos - 1
106 if (substring(line, end_pos, end_pos) == ' ') found_space <- TRUE
107 }
108 }
109
110 trimws(substring(line, start_pos, end_pos))
111 })
112 l_data[[length(l_data) + 1]] <- setNames(data, txt_headings)
113 }
114 }
115 }
116 }
117 }
118
119 ## join tables
120 df <- do.call(rbind, lapply(l_all, function(x) x$data))
121
122 ## convert table columns to numeric
123 for (n in names(df)) {
124 if (all(grepl("^(-|\\+)?((\\.?\\d+)|(\\d+\\.\\d+)|(\\d+\\.?)|(\\d*\\.*\\d+[E|e](-|\\+)\\d+))$", df[[n]]))) {
125 df[[n]] <- as.numeric(df[[n]])
126 }
127 }
128 df
129})
130
131df_all <- do.call(rbind, l.df)
132
133write.csv(df_all, 'table_out.csv', row.names = FALSE)
134
Input File Example