Adding Metadata to R Functions
This post describes three ways to tag functions with metadata.
Attributes
Using attributes is a simple way to add metadata to any object in R. Data frames and other objects already use attributes to hold metadata which are accessible via the attributes function.
1df <- data.frame(a = 1:5, b = letters[1:5])
2
3## retrieve attributes
4attributes(df)
5$names
6[1] "a" "b"
7
8$class
9[1] "data.frame"
10
11$row.names
12[1] 1 2 3 4 5
Metadata can be added by simply using the attributes function and retrieved using attr.
1f <- function(x, y) {
2 sum(x, y)
3}
4
5## add two new attributes
6attributes(f) <- c(attributes(f), f = "function: sum", param_count = 2)
1## list all attributes
2attributes(f)
3
4$srcref
5function(x, y) {
6 sum(x, y)
7}
8
9$f
10[1] "function: sum"
11
12$param_count
13[1] 2
1## retrieve a single attribute
2attr(f, "param_count")
3[1] 2
Metadata in comment
Metadata can be associated with any R object using comment. The comment is, in fact, simply an attribute with the limitation that it must be a character vector. Multiple items of metadata can be attached using the comment function.
1f <- function(x, y) {
2 sum(x, y)
3}
4
5comment(f) <- c(f = "function: sum", param_count = "2")
Once set, comment can be used to retrieve one of more items of metadata.
1comment(f)
2 f param_count
3"function: sum" "2"
4
5comment(f)[["param_count"]]
6[1] "2"
For a more complex data structure a json object can be attached. Note that in the example below, param_count is a numeric as opposed to a character type imposed when using comment to hold a vector.
1f <- function(x, y) {
2 sum(x, y)
3}
4
5comment(f) <- jsonlite::toJSON(list(f = "function: sum", param_count = 2), auto_unbox = TRUE)
1## retrieving metadata (comment)
2comment(f)
3{"f":"function: sum","param_count":2}
4
5jsonlite::fromJSON(comment(f))
6$f
7[1] "function: sum"
8
9$param_count
10[1] 2
Metadata in roxygen2
Metadata can be included in custom roxygen2 tags. Once added it can be retrieved programatically.
In order to demonstrate this method we need to build two packages - one for the custom tag and the other to demonstate its use in a function.
Package 1 - Custom roxygen2 tag
This package consists of a number of functions to define a new roclet, metadata which can be used to store metadata for a function. The R code is shown below.
1#' roclet to parse @metadata tag
2#'
3#' @import roxygen2
4#' @export
5metadata <- function() {
6 roxygen2::roclet("metadata")
7}
8
9
10#' @rdname metadata
11#' @importFrom roxygen2 tag_markdown
12#' @export
13roxy_tag_parse.roxy_tag_metadata <- function(x) {
14 roxygen2::tag_markdown(x)
15}
16
17
18#' @rdname metadata
19#' @importFrom roxygen2 rd_section
20#' @export
21roxy_tag_rd.roxy_tag_metadata <- function(x, base_path, env) {
22 roxygen2::rd_section('metadata', x$val)
23}
24
25#' @rdname metadata
26#' @export
27format.rd_section_metadata <- function(x, ...) {
28 paste0(
29 "\\section{Metadata}{\n",
30 x$value,
31 "}\n"
32 )
33}
34
35
36#' @rdname metadata
37#' @export
38roclet_process.roclet_metadata <- function(x, blocks, env, base_path) {
39 x
40}
41
42
43#' @rdname metadata
44#' @export
45roclet_output.roclet_metadata <- function(x, results, base_path, ...) {
46 x
47}
An explanation of extending roxygen2 can be found with the roxygen2 documentation.
After running roxygen2::roxygenize() to document and build the NAMESPACE file, this package can be built and installed.
Package 2 - testing
To test the roclet we need to build a second package consisting of a single function, my_function.
1#' my function
2#'
3#' my function to test metadata roclet
4#'
5#' @metadata \{"a":1, "b":"text string", "c": \[4, 5, 6\]\}
6#'
7#' @export
8my_function <- function(x) {
9 x
10}
This function contains the new roclet @metadata which holds a json-encoded string of parameters.
In addition to the function we need to add this line to the DESCRIPTION file:
1Roxygen: list(markdown = TRUE, roclets = c("namespace", "rd", "collate", "roxymeta::metadata"))
Once roxygen2::roxygenize() is run, the function documentation incudes the new tag:
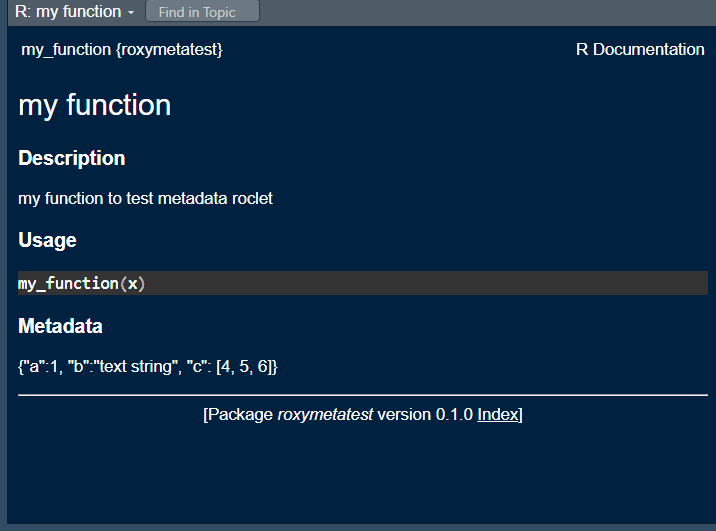
Function to retrieve metadata
Now that the metadata has been attached to a function it can be retrieved using the code below:
1#' extract parameters from roxygen tags
2#'
3#' extract parameters from roxygen tags
4#'
5#' @param n Namespace
6#' @param f Function
7#'
8#' @importFrom tools Rd_db
9#' @importFrom jsonlite fromJSON
10#'
11#' @export
12#'
13get_params <- function(n, f) {
14 db <- tools::Rd_db(n)
15 fn_rd <- db[[paste0(f, ".Rd")]]
16
17 ## get list of attributes
18 fn_attributes <- lapply(fn_rd, attributes)
19
20 ## get sections
21 fn_sections <- which(
22 sapply(fn_attributes, function(x) {
23 x$Rd_tag == "\\section"
24 })
25 )
26
27 ## get param section
28 fn_params <- which(
29 sapply(fn_rd[fn_sections], function(x) {
30 x[[1]] == "Metadata"
31 })
32 )
33
34 if (length(fn_params) > 0) {
35
36 input_tags <- fn_rd[[fn_sections[fn_params]]]
37 param_tag <- input_tags[[2]][[2]]
38 return(jsonlite::fromJSON(as.character(param_tag), simplifyVector = FALSE))
39
40 } else {
41
42 return(NULL)
43
44 }
45
46}
This function simply retrieves the Rd file and parses it, retrieving the json-encoded metadata and returning a list.
1out <- get_params('roxymetatest', 'my_function')
2out
3
4$a
5[1] 1
6
7$b
8[1] "text string"
9
10$c
11$c[[1]]
12[1] 4
13
14$c[[2]]
15[1] 5
16
17$c[[3]]
18[1] 6
19
This is a very basic example, simply demonstrating what can be done.
One use-case for this approach is for self-building shiny functions. Such a function could embed shiny widget details which are used to build interactive inputs to the function. The shiny widgets could therefore be built for the function prior to running the function itself.
code for this example is available in
https://github.com/harveyl888/roxymeta
https://github.com/harveyl888/roxymetatest
https://gist.github.com/harveyl888/1f043414a4102ed5f04e8ed22b73c939